By Yinjie Qiu
With about 450 species, fescues (Festuca L., Poaceae) are a large and diverse genus of perennial grasses (1). Fescue species are distributed mostly in temperate zones of both the northern and southern hemispheres, but most commonly are found in the northern hemisphere (2). Several of the fescue species are commonly used as turfgrass.
Based on various features, fine fescues are currently divided into two groups referred to as the F. rubra complex [includes F. rubra ssp. litoralis (slender creeping red fescue), F. rubra ssp. rubra (strong creeping red fescue), F. rubra ssp. fallax (Chewings fescue)] and the F. ovina complex [includes F. brevipila (hard fescue) and F. ovina (sheep fescue)] (3). While it is relatively easy to identify fine fescue species into their proper complex, it is challenging to differentiate species within the same complex. Species identification within the F. ovina complex relies heavily on leaf characteristics; however, abundant morphological and ecotype diversity within F. ovina makes species identification difficult (4). For example, leaf color can be a clue in plant species identification, but it is not a reliable indicator in the fescues. In the United States, the sheep fescue ecotypes tend to have a bluish-gray leaf color and the hard fescue ecotypes have a green leaf blade color (5) while in Europe, it is the opposite (6).
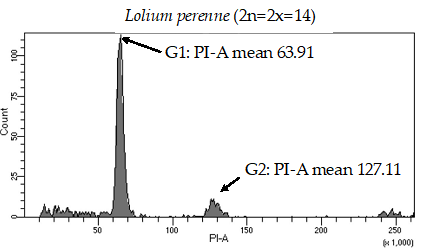
Historically, laser flow cytometry has been used to determine the number of chromosome sets in plant species (7). Flow cytometry is the measurement of cell characteristics, which can include cell size, cell count, cell cycle and more. This technique allows researchers to get highly specific information about individual cells. The nuclei extracted from fresh plant tissue are stained with propidium iodine, a DNA-staining chemical that emits fluorescence when lasers shoot on it. Samples are analyzed via a flow cytometer to quantify fluorescent intensity which correlates to the DNA content present in the nuclei. We have been using this methodology to effectively separate unknown fine fescue taxa by ploidy level (Figure 1; 8).
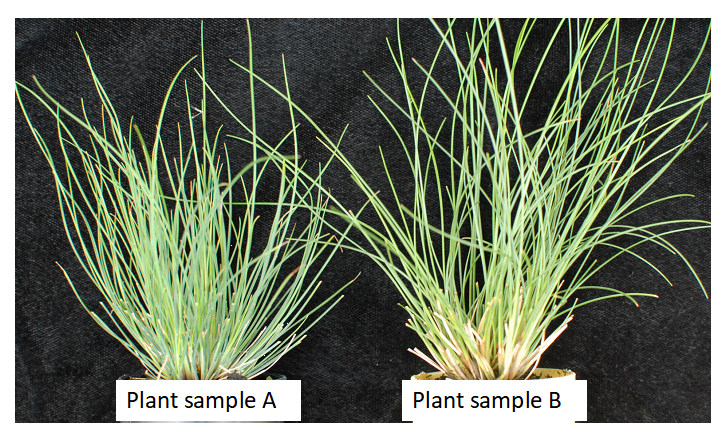
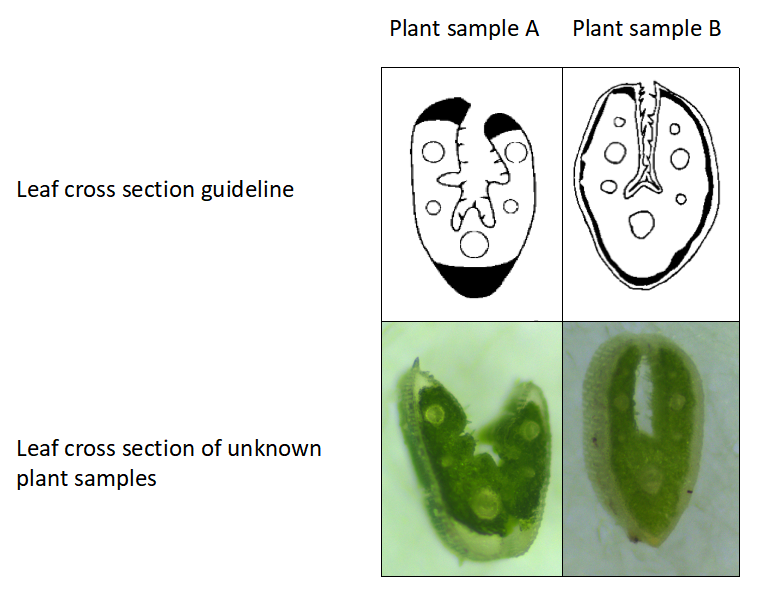
When fine fescue taxa have different morphology but the same ploidy level, leaf cross-sections can be used for further identification. For example, as shown in Figure 2, flow cytometry identified that plant A and B have the same chromosome numbers (diploids), but clearly they have different morphology. In a situation like this, we can use leaf-cross sections to confirm taxon identify. As shown in Figure 3, plant sample A has five veins while plant sample B has 7 veins. We are able to find their taxon based on the leaf cross section guideline (9,10).
By using flow cytometry, leaf cross section and previous marker development work (8), we will be able to more accurately identify fescue taxa. This effort will greatly facilitate fine fescue breeding and genetics work.
This research is supported under a grant, "Increasing Low-Input Turfgrass Adoption Through Breeding, Innovation, and Public Education", from the USDA-NIFA through the Specialty Crop Research Initiative.
References
- Clayton, W.D. and S.A. Renvoize, Genera graminum. Grasses of the world. Genera graminum. Grasses of the World., 1986. 13.
- Jenkin, T.J., Fescue Species (Festuca L.). In: Roemer, T. & W. Rudorf. Handbuch der Pflanzenzüchtung, 1959(4): p. 418–434.
- Ruemmele, B., L. Brilman, and D. Huff, Fine fescue germplasm diversity and vulnerability. Crop Science, 1995. 35(2): p. 313-316.
- Piper, C.V., North American species of Festuca. Vol. 10. 1906: US Government Printing Office.
- Beard, J.B., Turfgrass: Science and Culture. 1972.
- Hubbard, C.E., Grasses. A guide to their structure, identification, uses, and distribution in the British Isles. Grasses. A guide to their structure, identification, uses, and distribution in the British Isles., 1968.
- Huff, D.R. and A.J. Palazzo, Fine fescue species determination by laser flow cytometry. Crop Science, 1998. 38(2): p. 445-450.
- Qiu, Y., et al., Chloroplast Genome Sequencing and Comparative Analysis for Fine Fescue (Festuca L., Poaceae) Turfgrasses. bioRxiv, 2019: p. 708149.
- Wilkinson, M.J. and C.A. Stace, A new taxonomic treatment of the Festuca ovina L. aggregate (Poaceae) in the British Isles. Botanical Journal of the Linnean Society, 1991. 106(4): p. 347-397.
- Aiken, Susan G., and Laurie L. Consaul. Leaf cross sections and phytogeography: A potent combination for identifying members of Festuca subgg. Festuca and Leucopoa (Poaceae), occurring in North America. 82, no. 10 (1995): 1287-1299.