By Yinjie Qiu
Turfgrass grows in a complex environment that is full of stresses. For example, in the summertime, turfgrasses may be under abiotic stresses such as heat and drought stress. Some biotic stresses, such as summer patch and dollar spot disease, can occur at the same time. Situations like this cause stress for golf course superintendents and other turf managers. Some of these problems could be solved by cultural management methods; however, these methods can be labor intensive and expensive. Breeding turfgrass species with better stress resistance could solve these problems in more economical and ecosystem-friendly ways.
In nature, plants overcome abiotic and biotic stress by expressing stress-related genes and producing associated compounds. For instance, under heat stress, a plant produces a heat shock protein to protect itself, while under disease invasion, plants produce pathogenesis-related proteins to fight off disease. In turfgrasses, due to their outcrossing nature, even the seeds from the same seed lot could display a certain degree of genetic variation that leads to phenotypic variation under stressful environments. As such, discovering genes that are important for stress tolerance is crucial in developing improved turfgrass cultivars. In the past, discovering new genes in turfgrass species was difficult due to the complexity of the grass genome. This process has been simplified significantly with the development of new biotechnological tools.
In molecular biology, information from DNA is first transcribed into RNA and eventually made into functional proteins. In other words, RNA is an expression part for the plant genome, and RNA will only be transcribed when the stress situation triggers the transcription. For this reason, obtaining biological information from RNA is the most effective way to go for discovering genes underlying the biological process.
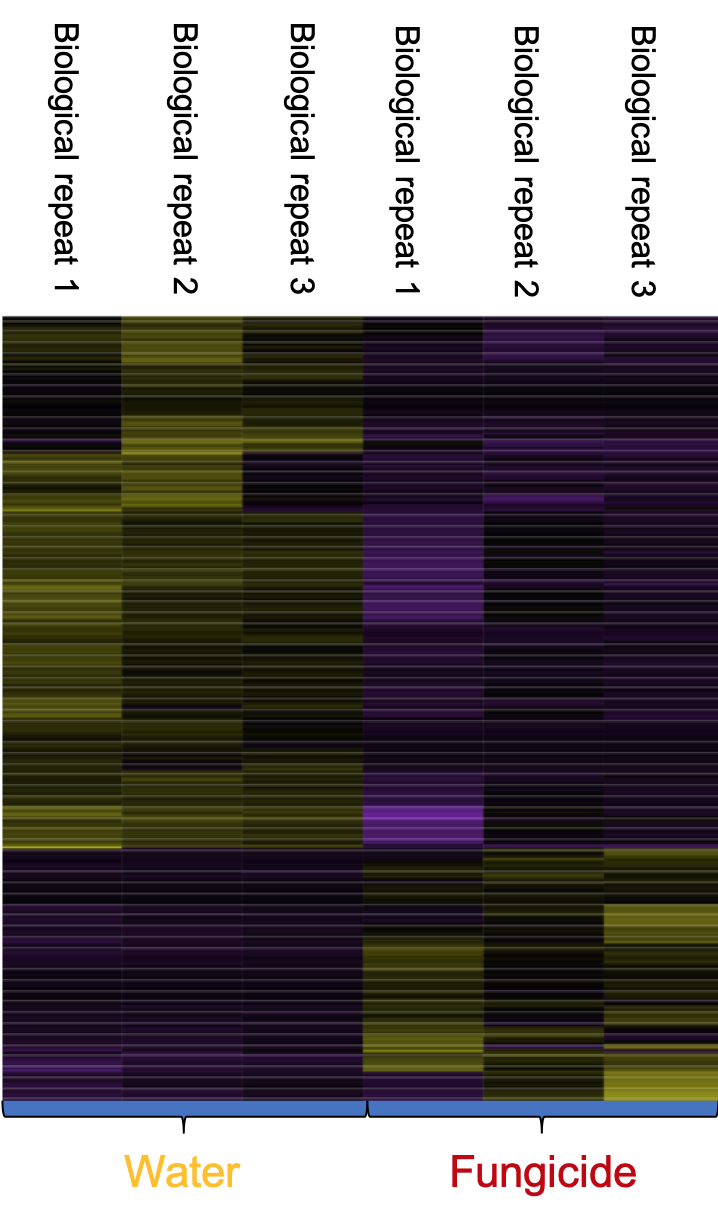
Decades ago, Illumina Inc. developed a high throughput sequencing platforms to sequence RNA (RNA-Seq). RNA sequencing produces millions of short sequences (100 nucleotide sequence each) in a relatively small amount of time. These sequences can be analyzed and reconstructed into genes. RNA sequencing also allows turfgrass scientists to look at which genes have been highly expressed in the stress environment and which genes expression have been turned off. In an example from our team’s research, we examined turfgrass response to fungicide application. Through RNA sequencing we were able to identify which genes were turned on and what genes have been turned off (Figure 1). Similarly, we can apply this method to look for genes related to disease response. Ideally, in a diverse population of plants, we could identify some candidate genes that can help turfgrass plants fight off pathogens such as pink snow mold. The identification of those genes will have the potential to help turfgrass breeders to breed cultivars that have better disease resistance.
As technology progresses, third generation sequencing (PacBio Sequencing) is now available for turfgrass scientists to further look at genes in turfgrass at a new level. See Table 1 for a comparison of the different eras of sequencing. As more and more gene sequences become available, breeding turfgrass for better stress tolerance using genetics tools will be more effective and we will be able to provide homeowners and turfgrass managers with better cultivars.
| Sequencing platform | First generation sequencing | Next generation sequencing | Third generation sequencing |
|---|---|---|---|
| Sequencing length in base pairs (bp) | 300-1000 | 50-300 | 30,000 |
| Sequencing throughput | Low | High | Medium |
| Sequencing accuracy | Accurate | Accurate | High error rate |