By Nicole Mihelich
Turfgrasses are diverse species that are of interest to researchers and consumers around the world. However, there has not been as much research conducted on turfgrasses compared to extensively studied model species and major world food crops. Investigating the developmental similarities between well-researched plants and turfgrasses can help translate insights to understand and improve turf.
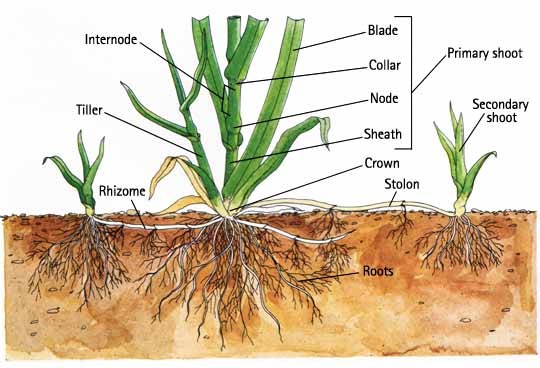
A major feature of many grasses is horizontal stem growth that enables the plants to spread and cover ground to create a lawn. Grass species all have vertically growing stems called tillers, but some species also have different types of specialized horizontal stems. Rhizomes are horizontal underground stems that are characteristic of some cool-season grasses, notably Kentucky bluegrass, strong creeping red fescue, and slender creeping red fescue. Stolons are similar to rhizomes as horizontal stems, but grow above the ground and are characteristic of the cool-season grass creeping bentgrass, as well as many warm-season grasses like centipede and St. Augustinegrass. Additionally, zoysia and Bermudagrass have both rhizomes and stolons. Tillers, rhizomes, and stolons all originate from axillary buds on the main stem/crown of the plant, yet their morphologies are distinct (Figure 1).
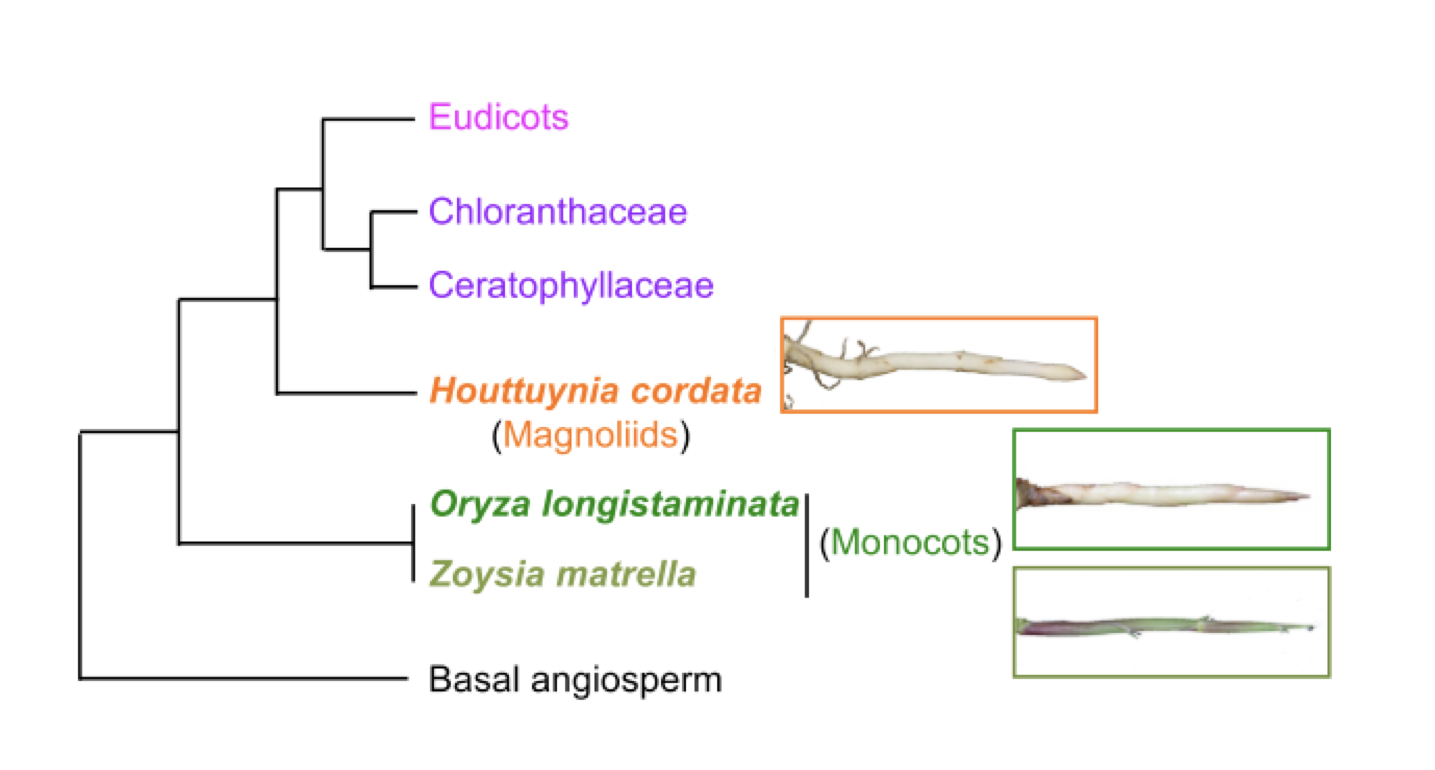
In 2003, a gene was found to control leaf patterning and stem growth in the famous model plant Arabidopsis thaliana and was called BLADE-ON-PETIOLE (BOP; Ha et al., 2003). Having evolved from a common ancestral gene, versions of this gene were later found in other plants like rice, and retained similar functions of regulating leaf and stem growth (Toriba et al., 2019). More recently, researchers took this investigation a step further in a paper by Toriba et al. (2020) entitled, “Suppression of Leaf Blade Development by BLADE- ON-PETIOLE Orthologs Is a Common Strategy for Underground Rhizome Growth”, published in the journal Current Biology. The authors achieved this by focusing on the rhizomes of Oryza longistaminata (a wild perennial relative of cultivated rice), the stolons of turfgrass Zoysia matrella, and the rhizomes of a broad-leaved species Houttuynia cordata (Figure 2).
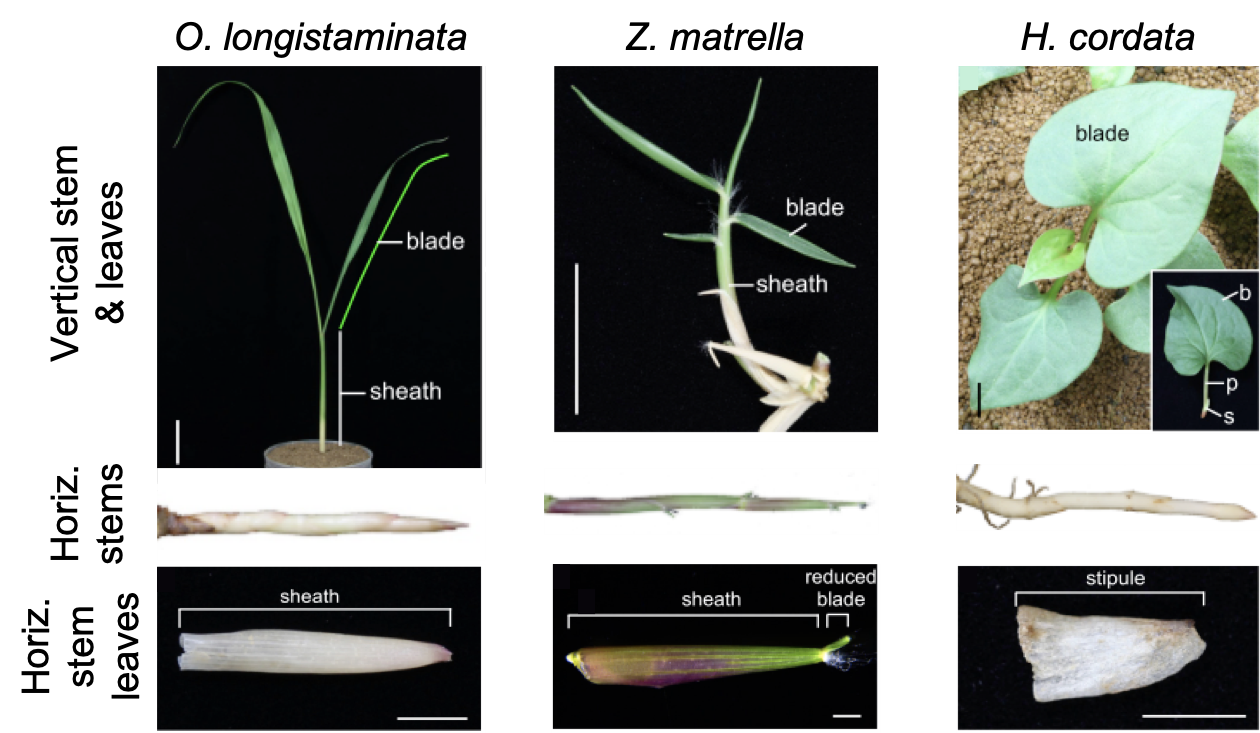
In grasses, aerial stems have leaves with a sheath growing around the stem, and a blade which extends away from the stem. Broad-leaved plants have similar patterns with a stipule and petiole at the base of the leaf, and the blade fanning out (Figure 3, Vertical stem & leaves). Rhizomes and stolons also have leaves, but Toriba et al. (2020) found that their leaves were either lacking blades, or the blades were extremely reduced. This led to the researchers’ first conclusion that blade formation is suppressed in rhizome and stolon leaves, which seems to be common across plants with horizontal stems.
Next, to investigate if BOP genes (those controlling leaf patterning and stem growth in Arabidopsis, as described earlier) are potentially involved in this observed blade suppression, Toriba et al. (2020) measured the expression levels and locations of expression of natural BOP genes. They found across three different plant species that BOP expression was upregulated and more widely expressed in rhizome and stolon leaves compared to aerial shoot leaves, suggesting that BOP genes may be important across rhizome- and stolon-producing plants.
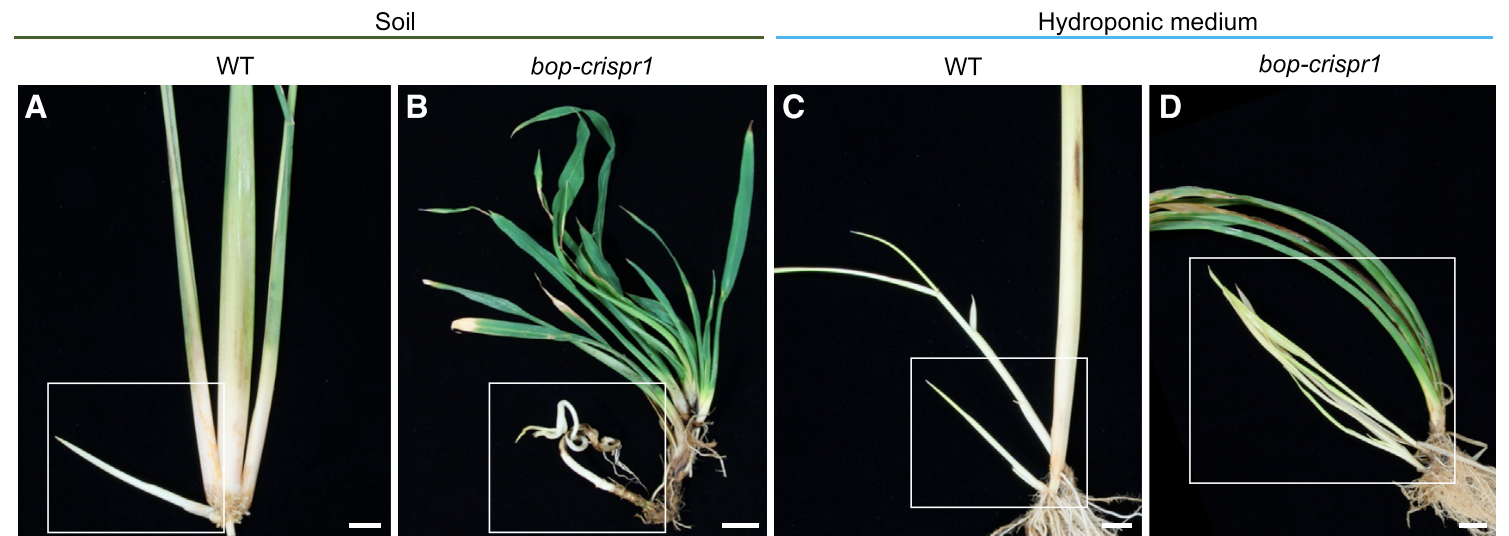
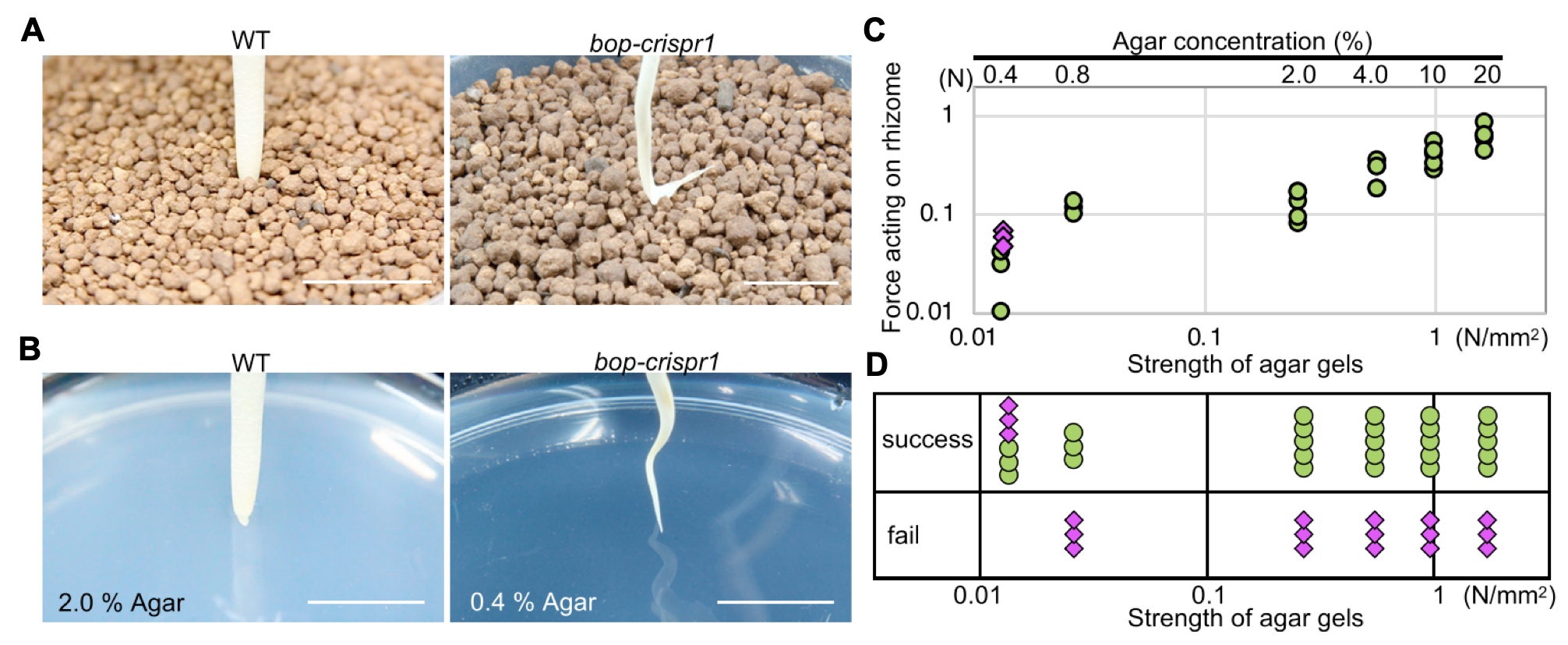
These expression studies are a great first step to investigate potential genes involved in horizontal stem growth, but they do not tell us how or if the gene is affecting the development of these stems and their ultimate form. Mutant plants that have non-functioning genes of interest can provide insights into what these genes are doing when they were properly functioning. To more concretely characterize the function of BOP genes in O. longistaminata rhizome development, Toriba et al. (2020) used CRISPR-Cas9 genome editing technology to disrupt the function of three BOP genes. They then compared the non-edited wild-type plant to the bop mutant plants grown in soil as well as in a hydroponic system. Wild-type (the ones that were not edited) plant tillers had distinct sheath and blades while rhizomes have virtually no sheaths. In contrast, the tillers and rhizomes of in the bop mutants had much more leaf blade growth than the wild-type (Figure 4). They observed that rhizomes of wild-type plants in soil grew straight, while bop mutants had very twisted and tangled rhizomes (Figure 4A-B). This suggested that the mutant rhizomes may be having difficulties growing in soil. In the hydroponic system where soil would not be a barrier to growth, both wild-type and bop mutant rhizomes grew straight (Figure 4C-D). This finding suggested that the predominantly sheath leaves of rhizomes may be important for the ability to grow underground.
To further test that rhizome sheath leaves are structurally important for underground growth, Toriba et al. (2020) measured the wild-type and mutant rhizomes’ ability to penetrate different surfaces, and how much force the rhizomes could withstand while doing so. First, manually forced rhizomes of the wild-type were found to easily penetrate the soil, while bop mutant rhizomes could not, and instead bent at the tip (Figure 5A). To better quantify these apparent differences in rhizome-tip stiffness, different concentrations of agar gels were used: the higher the concentration of agar, the more force was required to break through the surface. The wild-type rhizomes could break through the agar at all concentrations, while the bop mutant rhizomes could only penetrate the lowest agar concentration of 0.4% (Figure 5B-D). These experiments showed that the leaf sheath is necessary to create a stiff and sharp cone-shaped “drill” for successful rhizome growth underground. While these experiments were only conducted in O. longistaminata, these observations along with the previous experiments with two other species in the paper show that these mechanisms for rhizome growth may be common to other species. Since rhizome growth in compacted soils is often necessary for successful turfgrass stand development, the authors’ work can be an important resource for turfgrass scientists.
Research on rhizomes and stolons in O. longistaminata, Z. matrella, and H. cordata has important insights that can improve turfgrass. As mentioned above, many turfgrass species have rhizomes and/or stolons that can play a major role in horizontal spread and rapid lawn establishment. This advantage is applicable in direct-seeded/sprigged lawns, but may be especially important in sod production because sod requires a thick mat of interlocking material that can hold together during harvest and transplanting. Research on turfgrasses improving rhizome and/or stolon growth can lead to more rapid lawn establishment and competitiveness against weeds, potentially leading to less erosion, run-off and herbicide use. Understanding the mechanisms of stronger rhizome tip growth could also lead to improvement of rhizomatous turfgrasses that fare better in compacted soils. This research can even extend to other horticultural research as well, for instance, being able to “turn off” rhizome production in some ornamentals to prevent invasive spreading in a landscape. Although much research is still needed to translate insights from model species, this research is a crucial step in advancing knowledge for breeding efforts of turfgrass and beyond with horizontal stem traits.
References
Ha, C. M., Kim, G. T., Kim, B. C., Jun, J. H., Soh, M. S., Ueno, Y., ... & Nam, H. G. (2003). The BLADE-ON-PETIOLE 1 gene controls leaf pattern formation through the modulation of meristematic activity in Arabidopsis. Development, 130(1), 161-172.
Toriba, T., Tokunaga, H., Shiga, T., Nie, F., Naramoto, S., Honda, E., ... & Kyozuka, J. (2019). BLADE-ON-PETIOLE genes temporally and developmentally regulate the sheath to blade ratio of rice leaves. Nature communications, 10(1), 1-13.
Toriba, T., Tokunaga, H., Nagasawa, K., Nie, F., Yoshida, A., & Kyozuka, J. (2020). Suppression of leaf blade development by BLADE-ON-PETIOLE orthologs is a common strategy for underground rhizome growth. Current Biology, 30(3), 509-516.
Zeng, L., Zhang, Q., Sun, R., Kong, H., Zhang, N., & Ma, H. (2014). Resolution of deep angiosperm phylogeny using conserved nuclear genes and estimates of early divergence times. Nature communications, 5(1), 1-12.